17 April 2014 - New Motif Displays for Transcription Factor ChIP-seq Track and New Genome Segmentations from ENCODE
The latest Transcription Factor ChIP-seq
track has been enhanced with the display of Factorbook motifs. Within a cluster, a green
highlight indicates the highest scoring site of a Factorbook-identified canonical motif for the
corresponding factor. Along with the ability to suppress motif highlights and cell abbreviations,
the track configuration page now also enables the filtering of factors.
The newly added
Genome Segmentations from ENCODE tracks display multivariate genome-segmentation performed on
six human cell types
(GM12878, K562, H1-hESC, HeLa-S3, HepG2, and HUVEC), integrating ChIP-seq data for
eight chromatin marks, RNA Polymerase II, the CTCF transcription factor and input data. In total,
twenty-five states were used to segment the genome, and these states were then grouped and colored to
highlight predicted functional elements. These Genome Segmentations are the same data as
found in the Analysis Working Group Hub, but now hosted natively in the Browser with enhanced
filtering capability where desired segmented states can be selected using the
'Filter by Segment Type' control on the track configuration page.
12 Sept 2013 - New UDR ENCODE Download Method Available
The UCSC Genome Browser is pleased to offer a new download protocol to use when downloading
large sets of files from our download servers: UDR (UDT Enabled Rsync). UDR utilizes rsync
as the transport mechanism, but sends the data over the UDT protocol, which enables huge
amounts of data to be downloaded efficiently over long distances.
Remember that we now have two identical download servers to better serve your needs. You can use either one:
http://hgdownload.cse.ucsc.edu
http://hgdownload-sd.sdsc.edu
The Background:
Typical TCP-based protocols like http, ftp and rsync have a problem in that the further
away the download source is from you, the slower the speed becomes. Protocols like UDT/UDR
allow for many UDP packets to be sent in batch, thus allowing for much higher transmit speeds
over long distances. UDR will be especially useful for users who are downloading from places
that are far away from California. The US East Coat and the international community will likely
see much higher download speeds by using UDR rather than rsync, http or ftp.
Getting UDR & Setting it up on your System:
It should be noted that UDR is not written or managed by UCSC, it was written by the
Laboratory for Advanced Computing at the University of Chicago. It has been tested to work
under Linux, FreeBSD and Mac OSX, but may work under other UNIX variants. The source code can
be obtained here, through GitHub:
https://github.com/LabAdvComp/UDR
If you need help building the UDR binaries or have questions about how UDR functions,
please read the documentation on the GitHub page, and if necessary, contact the UDR authors
via the GitHub page. We recommend reading the documentation on the UDR GitHub page to better
understand how UDR works. UDR is written in C++. UDR is Open Source and is released under the
Apache 2.0 License. You must first have rsync installed on your system.
For your convenience, we are offering a binary distribution of UDR for Red Hat Enterprise
Linux 6.x (or variants such as CentOS 6 or Scientific Linux 6). You'll find both a 64-bit
and 32-bit rpm here:
http://hgdownload.cse.ucsc.edu/admin/udr
Using UDR to Download ENCODE Data from the UCSC Genome Browser Download Server(s):
Once you have a working UDR binary, either by building from source or by installing the rpm
(if you are using RHEL 6.x or other variant), you can download files from either of our our
download servers in a very similar fashion to rsync. For example, using rsync, you may want
to download all of the ENCODE information for the mm9 database using the following command:
$ rsync -avP rsync://hgdownload.soe.ucsc.edu/goldenPath/mm9/encodeDCC/ /my/local/mm9/
Using UDR is very similar. The UDR syntax for downloading the same data would be:
$udr rsync -avP hgdownload.cse.ucsc.edu::goldenPath/mm9/encodeDCC/ /my/local/mm9/
If you installed the rpm, use the 'man udr' command for more information via the man page;
if you installed from source please refer to the UDR GitHub page for more details on the
capabilities of UDR and how to use it.
Firewall Considerations:
UDR establishes connections on TCP/9000, then transmits the data stream over UDP/9000-9100.
Your institution may need to modify its firewall rules to allow inbound and outbound ports
TCP/9000 and UDP/9000-9100 from either of the two download machines.
If you decide to install and use UDR, we hope that you experience greatly increased download
speeds. If you have difficulties installing UDR on your system, please contact the Laboratory
for Advanced Computing through their gitHub page: https://github.com/LabAdvComp/UDR.
25 July 2013 - BLUEPRINT Epigenome Data Hub and Quick Reference PDF Now Available
We are pleased to announce the addition of the
BLUEPRINT Epigenomics Data Hub on the UCSC Genome Browser through our
Public Hubs
function. All data were produced and processed by the European
BLUEPRINT Epigenome project,
aimed to generate 100 reference epigenomes from distinct types of haematopoietic cells and
their malignant leukaemic counterparts. Please send any data related questions to
blueprint-info@ebi.ac.uk. The BLUEPRINT Hub currently contains 8 DNase-seq, 48 DNA methylation,
170 Histone modification, and 24 RNA-seq tracks, focused on displaying variation in human
monocyte and neutrophil cells from 7 adult blood and 4 cord blood samples. Future releases
of data for additional samples are planned at a regular basis.
Also the ENCODE Quick Reference Card is now available in
PDF courtesy of OpenHelix on the
ENCODE Education and Outreach page.
28 May 2013 - ENCODE portal changes: New Link to NHGRI Tutorials, New External
Software Tools Page, Updates to Publications including New 2013 Consortium Papers Section
The ENCODE portal was updated to include informative new and expanded pages.
A new link to the NHGRI ENCODE Tutorials was added to the
ENCODE Education and Outreach page.
A new External Software Tools
page was added to expand the existing ENCODE Software Tools
resources page. The Publications page was expanded to include a new
2013 Consortium-funded section
and updated with several new references as well as the non-ENCODE Publications page.
Lastly other pages were updated with new information including the ENCODE Analysis Software page, and Quality Metrics page.
13 May 2013 - Uniform Peaks of Transcription Factor ChIP-seq from ENCODE/Analysis
UCSC has released a new browser track containing 690 datasets of transcription factor ChIP-seq
peaks based on data from all five ENCODE TFBS ChIP-seq production groups from the project
inception in 2007 through the ENCODE March 2012 data freeze.
The track covers 161 unique regulatory factors (generic and sequence-specific factors),
spanning 91 human cell types, some under various treatment conditions.
Browser track:
Transcription Factor ChIP-seq Uniform Uniform Peaks from ENCODE/Analysis
File downloads:
Directory
File selection tool
This track represents peak calls (regions of enrichment) generated by the ENCODE Analysis Working
Group (AWG) using the uniform processing pipeline developed for the ENCODE Integrative Analysis
effort and published in a set of coordinated papers in September 2012. Peak calls from that
effort (based on datasets from the January 2011 ENCODE data freeze) are available at the
ENCODE Analysis Data Hub.
The new Uniform TFBS track at UCSC includes newer data, slightly modified processing methods,
and improved metadata. Quality metrics are included in metadata, with detailed metrics in a
quality spreadsheet linked to the track description.
Browser users will see the uniform peaks first when using track search for TFBS, and
this track is now the default track shown when the ENCODE TF Binding menu item is
selected in the browser.
The primary and lab-processed data (along with methods descriptions, credits and references)
on which this track is based are available in the following ENCODE tracks: HAIB TFBS, SYDH TFBS,
UChicago TFBS, UTA TFBS, UW CTCF Binding. Many thanks to Anshul Kundaje of the ENCODE AWG
for providing the uniform peaks data, description, and quality spreadsheets.
7 April 2013 - User Resources and Frequently Asked Questions page added to website
This new page provides links to
ENCODE informational material and tools at the NHGRI,
GEO, UCSC, and Nature, together with links to some of the most useful pages at encodeproject.org.
It also includes a helpful FAQ section culled from ENCODE questions received on the ENCODE and
Genome Browser mailing lists.
25 January 2013 - Uniform DNaseI Hypersensitivity sites from ENCODE Analysis Working Group
The ENCODE Analysis Working Group (AWG) has performed uniform processing
on datasets produced by multiple data production groups in the ENCODE
Consortium.
The AWG uniform datasets are used in downstream analysis pipelines by
members of the ENCODE Consortium and are one of the primary sources of
data referenced in the 2012 ENCODE integrative analysis.
UCSC has released a new browser track and a track update based on the AWG uniform processing of
ENCODE DNaseI data:
DNaseI Hypersensitivity Uniform Peaks from ENCODE/Analysis:
This track displays the uniform set of open chromatin elements in 125 ENCODE
cell types on a per-cell type basis.
Digital DNaseI Hypersensitivity Clusters in 125 cell types from ENCODE:
This track displays clusters of Uniform DNaseI Hypersensitive sites across the cell types assayed.
This is the second release of the Dnase Clusters track, which is part of the ENCODE
Regulation supertrack in the Genome Browser.
It contains 51 additional cell type + treatment combinations. It differs from the
previous track by including data from multiple ENCODE groups that have been uniformly processed,
with replicates merged.
The previous track is available on the UCSC preview browser as
DNase Clusters V1.
The uniform elements in these tracks are based on DNase-seq data produced by the "Open Chromatin"
(Duke/UNC/UT-A) and University of Washington (UW) ENCODE groups from the project inception in 2007
through the ENCODE January 2011 data freeze.
The primary and lab-processed data (along with methods descriptions,
credits and references) on which these tracks are based are available in
the following ENCODE tracks:
ENCODE Duke DNaseI,
ENCODE UW DnaseI HS.
28 September 2012 - New ENCODE on-line tutorial and Quick Reference Cards
To supplement the existing ENCODE Foundations tutorial, OpenHelix has developed a second
tutorial describing newer ENCODE data and access tools. Slides for this tutorial are freely available through March 2013 from the
website at: http://openhelix.com/ENCODE2.
Also available free (up to 30 cards shipped free in the US) is a Quick Reference Card that
provides a handy summary of access features. For more information see
the OpenHelix announcement.
27 September 2012 - ENCODE data releases: UNC/BSU ProtGenc, UNC FAIRE (Rel 2), HAIB TFBS (Rel 3), CSHL Long RNA-seq (Rel 3)
One new track and three track updates were released on the human hg19 browser:
Proteogenomics Hg19 and GENCODE Mapping from ENCODE/Univ. North Carolina/Boise State Univ:
This track displays mass spectrometry data that have been matched to genomic sequences
in GM12878, H1-hESC, H1-neuron, and K562 cell types. Peptides were mapped to an in silico
translation and proteolytic digestion of the whole human genome (UCSC Hg19), and the GENCODE
translation of protein-coding transcripts database. The track can be used to identify which parts
of the genome are translated into proteins, to verify which transcripts discovered by other ENCODE
experiments are protein-coding, to reveal new genes and/or splice variants and proteins with
post-translational modifications (PTM). Of particular interest is the possibility of uncovering the
translation of small open reading frames (ORFs), antisense transcripts, or protein-coding regions
that have been annotated as introns previously.
Open Chromatin by FAIRE from ENCODE/OpenChrom(UNC Chapel Hill) (Release 2):
FAIRE (Formaldelhyde-Assisted Isolation of Regulatory Elements) is a method to isolate and identify
nucleosome-depleted regions of the genome. Release 2 of this track contains 12 new experiments,
including 11 new cell lines.
Transcription Factor Binding Sites by ChIP-seq from ENCODE/HAIB (Release 3):
This release contains 110 new experiments including 3 new cell lines.
There were also data corrections (see Release Notes section for details).
Long RNA-seq from ENCODE/Cold Spring Harbor Lab (Release 3):
This release contains additional files for many experiments including
GENCODE V10 referenced Transcripts, Genes and Exons (previous datasets
referenced GENCODE V7). Complete details are in the track Release Notes.
25 September 2012 - Mouse ENCODE data releases: UW DNaseI DGF, UW DNaseI HS (Rel 2), LICR Histone (Rel 3), CSHL Long RNA-seq (Rel 3)
One new track and three track updates were released on the Mouse mm9 browser:
DNaseI Digital Genomic Footprinting from ENCODE/University of Washington:
This track contains deep sequencing DNase data that can be used to identify sites where regulatory
factors bind to the genome (footprints). Footprinting is a technique used to define the DNA
sequences that interact with and bind DNA-binding proteins, such as transcription factors,
zinc-finger proteins, hormone-receptor complexes, and other chromatin-modulating factors like CTCF.
The technique depends upon the strength and tight nature of protein-DNA interactions. This track
contains a total of 22 DGF experiments, covering 20 mouse cell types and tissues.
DNaseI Hypersensitivity by Digital DNaseI from ENCODE/University of Washington (Release 2):
Adds 32 new experiments including 22 new cell lines and 4 new treatments.
Histone Modifications by ChIP-seq from ENCODE/LICR (Release 3):
Adds 31 new experiments for a total of 130 experiments.
Long RNA-seq from ENCODE/Cold Spring Harbor Lab (Release 3):
This release adds data for bladder, cerebellum, CNS, cortex, frontal lobe, limb, liver, placenta,
and whole brain. The samples for CNS, liver, limb and whole brain vary over age (developmental
stage). This release also contains replacement BAM files for the previous ones had the second read
reverse complemented.
5 September 2012 - ENCODE results published in Nature, Science and other journals
The results of the ENCODE project were published today in a coordinated set of 30 papers published in multiple journals.
These publications are the result of cross-consortium integrative analysis, covering more than 4
million regulatory regions in the human genome mapped as part of ENCODE. The coordinated publication
set includes one main integrative paper and five other papers in the journal Nature; 18 papers in
Genome Research; and six papers in Genome Biology. The ENCODE data are so complex that the three
journals have developed a pioneering way to present the information in an integrated form they term
"threads." Since the same topics were addressed in different ways in different papers, the
Nature ENCODE website
was developed to allow readers to follow a topic through
all of the papers in the ENCODE publication set. In addition to these publications, six review
articles are being published in the Journal of Biological Chemistry, and other affiliated papers in
Science, Cell, and other journals. The new
Integrative Analysis page on this portal provides links
and descriptive material for these publications and related analysis resources.
28 August 2012 - ENCODE data releases: Updates to UW DNaseI DGF, Duke DNaseI HS, Broad Histone, GIS RNA PET, SYDH TFBS, UTA TFBS
DNaseI Digital Genomic Footprinting from ENCODE/University of Washington (Release 4)
Adds 10 new experiments across 8 cell lines.
Open Chromatin by DNaseI HS from ENCODE/OpenChrom(Duke University) (Release 3):
Adds 27 new experiments including 18 new cell lines.
Histone Modifications by ChIP-seq from ENCODE/Broad Institute (Release 3):
Adds 83 new experiments including 6 new cell lines and 25 new antibodies.
RNA Sub-cellular Localization by Paired-end diTag Sequencing from ENCODE/GIS (Release 2):
Adds data for 4 additional cell lines (A549 lung carcinoma, SK-N-SH neuroblastoma, IMR90 lung fibroblast, and MCF-7 breast carcinoma).
Transcription Factor Binding Sites by ChIP-seq from ENCODE/Stanford/Yale/USC/Harvard (Release 3):
Adds 37 new experiments including 1 new cell line (SK-N-SH neuroblastoma) and 7 new antibodies.
Open Chromatin TFBS by ChIP-seq from ENCODE/Open Chrom(UT Austin) (Release 2) :
Adds 34 new experiments including 17 new cell lines.
21 August 2012 - Mouse ENCODE data releases: DNaseI HS from Penn State, Histone ChIP-seq from Penn State (Rel 2) and Stanford/Yale (Rel 2), TFBS ChIP-seq from Stanford/Yale (Rel 4), and Penn State (Rel 2)
DNaseI Hypersensitivity by Digital DNaseI from ENCODE/PSU
This track provides maps of DNaseI sensitivity in G1E (GATA- erythroid progenitor cells) untreated and differentiated via estradiol treatment. DNaseI has long been used to map general chromatin accessibility, and DNaseI hypersensitivity is a universal feature of active cis-regulatory sequences.
Histone Modifications by ChIP-seq from ENCODE/PSU (Release 2):
This release adds one new experiment (H3K4me1 in G1E cells).
Histone Modifications by ChIP-seq from ENCODE/SYDH (Release 2):
This release adds 12 new experiments, including 1 new cell line (E14 ES cells) and 5 new antibodies.
Transcription Factor Binding Sites by ChIP-seq from ENCODE/PSU (Release 2):
This release adds 14 new experiments.
Transcription Factor Binding Sites by ChIP-seq from ENCODE/Stanford/Yale (Release 4):
This release adds 22 experiments including 12 new antibodies.
9 August 2012 - Experiment Lists updated
The ENCODE DCC has updated the spreadsheets detailing ENCODE submissions
to reflect data status in the ENCODE Y5Q3 (July 1) timeframe.
These spreadsheets include all datasets that have been successfully
submitted to the DCC. Datasets having status 'released' are available on
the UCSC public site; others have not completed quality review, but are
available provisionally on the UCSC preview site.
These pages can be accessed from the 'Experiment List' links in the left menu.
3 August 2012 - Human ENCODE data releases: UW DNaseI HS (Rel 6), HAIB Methyl RRBS (Rel 3), FSU Repli-chip (Rel 2), UW Affy Exon (Rel 4)
DNaseI Hypersensitivity by Digital DNaseI from ENCODE/University of
Washington (Release 6):
Includes 11 new experiments across 12 new cell lines: bone marrow HS27a, bone marrow HS5, bone marrow MSC, CD4+ Naive Wb11970640, CD4+ Naive Wb78495824, Th17, Th1 Wb33676984, Th1 Wb54553204, Th2 Wb33676984, Th2 Wb54553204, Treg Wb78495824, Treg Wb83319432.
DNA Methylation by Reduced Representation Bisulfite Seq from ENCODE/HudsonAlpha (Release 3):
Adds the MCF-7 cell line with shRNA knockdowns obtained from the Crawford Lab at Duke University.
Replication Timing by Repli-chip from ENCODE/FSU (Release 2):
Adds 6 more data sets including additional replicates for H1-hESC and H7-hESC and all new data for the iPS skin fibroblast bio samples.
Affymetrix Exon Array from ENCODE/University of Washington (Release 4) (2012-07-31):
Adds in H7-hESC cells that have been differentiated with different time courses.
25 July 2012 - Mouse ENCODE data releases: UW RNA-seq, FSU Repli-chip, CSHL Long RNA-seq (Release 2)
Replication Timing by Repli-chip from ENCODE/FSU:
shows genome-wide assessment of DNA replication timing using NimbleGen tiling CGH microarrays. Each experiment represents the relative enrichment of early vs. late S-phase nascent strands in a given cell line, with data represented as a loess-smoothed function of individual timing values at probes spaced at even intervals across the genome. Regions with high values indicate domains of early replication where initiation occurs earlier in S-phase or early in a higher proportion of cells.
RNA-seq from ENCODE/UW:
shows RNA-seq measured genome-wide in 25 mouse tissues and cell lines. Poly-A selected mRNA was used as the source for transcriptome profiling of tissues and cell types that also had corresponding DNase I hypersensitive profiles.
Long RNA-seq from ENCODE/Cold Spring Harbor Lab (Release 2):
includes a new browser view (Splice Junctions) and 6 new analysis files per experiment (de novo exons, genes and transcripts with expression levels defined by Cufflinks and Flux Capacitor, and expression levels for Ensembl and Cufflinks models at each level).
16 July 2012 - Human ENCODE data releases: CSHL Small RNA-seq (Release 3), UCSC Regulation Transcription Track (Release 2), UW Affy Exon (Release 3), UMass 5C (Release 2)
Small RNA-seq from ENCODE/Cold Spring Harbor Lab (Release 3) :
adds 53 additional experiments, with twenty-two new cell lines represented: CD20+, CD34+_Mobilized, HAoAF, HAoEC, HCH, HFDPC, HMEpC, hMSC-AT, hMSC-UC, HOB, HPC-PL, HPIEpC, HSaVEC, HVMF, HWP, IMR90, Monocytes-CD14+, NHDF, NHEM.f_M2, NHEM_M2, SkMC, SK-N-SH. There are also two new variations in protocol (CIP-TAP and untreated) used to create different RNA populations.
UCSC Integrated Regulation: Transcription Levels Assayed by RNA-seq on 9 Cell Lines from ENCODE:
includes transcription levels for 2 additional cell lines: HeLa-S3 and HepG2.
Affymetrix Exon Array from ENCODE/University of Washington (Release 3):
revokes previously released K562 zinc-finger experiments due to a data merging issue.
Chromatin Interactions by 5C from ENCODE/Dekker Univ. Mass. (Release 2):
contains reanalyzed peak files and more complete raw data.
02 July 2012 - Quality metrics for human ENCODE data
The ENCODE Consortium analysis working group has analyzed the quality of the data produced using a variety of metrics. The ENCODE portal
Quality Metrics page provides
spreadsheets showing quality metrics for human ENCODE datasets, along with descriptions of what the metrics are and what they appear to measure. Spreadsheets are provided for DNase-seq, FAIRE-seq, Transcription Factor ChIP-seq, Histone ChIP-seq, and ChIP Controls. This page is linked from the related
Data Standards page.
Many of the software tools used for quality metrics, along with their citations, can be found on the portal
Software Tools page.
26 Jun 2012 - Human ENCODE data releases: GENCODE Genes V12, CSHL Long RNA-seq (Release 2), SYDH Histone (Rel 3), UW DNaseI HS (Rel 5)
Gene Annotations from ENCODE/GENCODE Version 12:
This version is based on the Ensembl 67 (May 2012) and Vega 47 (April 2012) gene predictions and curation.
Long RNA-seq from ENCODE/Cold Spring Harbor Lab (Release 2):
includes 25 new cell lines: CD20+, CD34+_Mobilized, HAoAF, HAoEC, HCH, HFDPC, HMEpC, hMNC-CB, hMNC-PB, hMSC-AT, hMSC-BM, hMSC-UC, HOB, HPC-PL, HPIEpC, HSaVEC, HVMF, HWP, IMR90, Monocytes-CD14+, NHDF, NHEM.f_M2, NHEM_M2, SkMC, and SK-N-SH. The downloadable elements files (Transcripts, Genes and Exons) in this release were generated using GENCODE V10, while the older datasets were generated using GENCODE V7.
Histone Modifications by ChIP-seq from ENCODE/Stanford/Yale/USC/Harvard (Release 3):
adds 9 new experiments for the MCF-7, HCT-116 and PANC-1 cell lines.
DNaseI Hypersensitivity by Digital DNaseI from ENCODE/University of Washington (Release 5):
adds 11 experiments across 9 new cell lines.
25 Jun 2012 - Mouse ENCODE data releases: Stan/Yale RNA-seq, Stan/Yale TFBS (Release 3)
:
One new track and one track update were released on the mm9 genome browser:
RNA-seq from ENCODE/Stanford:
This track shows enrichment of RNA sequence tags mapped to the mouse genome generated by high throughput sequencing in CH12, MEL, and ES cells.
Transcription Factor Binding Sites by ChIP-seq from ENCODE/Stanford/Yale (Release 3):
adds 17 new experiments, for a total of 54 experiments, studying 26 factors in CH12, MEL and ES cells.
15 Jun 2012 - Mouse Experiment Matrix web tool
The ENCODE DCC at UCSC has released an 'Experiment Matrix' for Mouse ENCODE data on the portal.
The three web pages in this application
(Experiment Matrix,
Experiment Summary, and
ChIP-seq Experiment Matrix)
provide an up-to-date view of the publicly
available Mouse ENCODE data, along with an interface for selecting
experiments for viewing in the browser or downloading as files for
analysis. Each page has a file/track selector. Clicking an experiment
item produces a search window with resulting files or tracks listed.
The pages are best viewed in Firefox, Chrome, and Safari browsers, and
zoomed out as far as readability on your screen allows.
31 May 2012 - Human ENCODE data releases: GIS ChIA-PET, SYDH TFBS (Release 2), RIKEN CAGE Loc (Release 3)
Chromatin Interaction Analysis Paired-End Tags (ChIA-PET) from ENCODE/GIS-Ruan:
This track shows the locations of protein factor mediated chromatin interactions determined
by ChIA-PET data in five different human cancer cell lines (K562 (chronic myeloid leukemia),
HCT116 (colorectal cancer), HeLa-S3 (cervical cancer), MCF-7 (breast cancer),
and NB4 (promyelocytic)). The protein factors displayed in the track include
estrogen receptor alpha, RNA polII, and CTCF.
Chromatin interaction analysis with paired-end tag sequencing (ChIA-PET) is a global
de novo high-throughput method for characterizing the 3-dimensional structure of chromatin
in the nucleus. A chromatin interaction is defined as the association of two regions
of the genome that are far apart in terms of genomic distance, but are spatially
proximate to each other in the 3-dimensional cellular nucleus.
Transcription Factor Binding Sites by ChIP-seq from ENCODE/Stanford/Yale/USC/Harvard (Release 2):
contains 79 new experiments.
RNA Subcellular CAGE Localization from ENCODE/RIKEN (Release 3):
includes viewable Transcription Start Sites determined by Hidden Markov Models. It contains the following new cell lines and tissues: CD20+, CD34+ Mobilized, HAoAF, HAoEC, HCH, HFDPC, HMEpC, hMSC-AT, hMSC-BM, hMSC-UC, HOB, HPC-PL, HPIEpC, HSaVEC, HVMF, HWP, IMR90, Monocytes-CD14+, NHDF, NHEM.f_M2, NHEM_M2, SkMC, and SK-N-SH.
25 May 2012 - Mouse ENCODE data releases: Caltech RNA-seq, PSU Histone, LICR RNA-seq (Release 2)
Two new tracks and one track update were released on the mm9 genome browser:
RNA-seq from ENCODE/Caltech:
This track shows transcriptome measurements in 10T1/2 fibroblasts and C2C12 myoblasts,
and these same cell types treated with differentiation media to produce fibrocytes and myocytes.
RNA-seq was performed on total cellular polyA+ RNA with 100bp paired end reads aligned to the genome.
Histone Modifications by ChIP-seq from ENCODE/PSU:
This track displays levels of three histone modifications - H3K4me1 (indicative of active chromatin and enhancers), H3K4me3 (enriched at active promoters), and H3K27me3 (associated with some silenced genes), in 5 blood-derived primary cells and cell lines.
RNA-seq from ENCODE/LICR (Release 2):
This release adds RNA-seq data for 12 additional primary cell types, bringing the total for this track to 22.
8 May 2012 - ENCODE Experiment Matrix web tool
The ENCODE DCC at UCSC is pleased to announce the release of a new
web tool for accessing ENCODE data.
The new
Experiment Matrix
link on the ENCODE portal
leads to a set of three web pages that provide an up-to-date view of the
breadth of human ENCODE data available, along with an interface for
selecting experiments for viewing in the browser or downloading as
files for analysis. (Note: the related
Experiment List
(previously 'Data Summary') spreadsheets have been updated to reflect newer status).
The main Experiment Matrix page shows the number of experiments for
each cell type/assay pairing. The related
ChIP-seq Experiment Matrix
provides a view of the transcription factor and histone modification
datasets, showing experiments by cell type and antibody target. The
Experiment Summary
lists the number of experiments by assay type
alone and includes annotations that are cell-type independent
(annotations on the reference genome). Each page has a file/track
selector. Clicking an experiment item produces a search window with
resulting files or tracks listed.
These pages are best viewed in FireFox, Chrome, and Safari browsers, and
zoomed out as far as readability on your screen allows.
Special thanks to Katrina Learned and Steve Heitner for careful QA review and testing,
to the Genome Browser build & release group for shepherding the new program
to the public site, and to the DCC wranglers for the high quality
metadata curation that supports this new access tool.
20 Apr 2012 - Human ENCODE data releases: UMass 5C, ENCODE Pilot Regions, FSU Repli-chip and 11 track updates
Three new tracks and 11 track updates were released on the hg19 genome browser:
Chromatin Interactions by 5C from ENCODE/Univ. Mass(Dekker):
This track contains chromatin interaction data generated using the 5C
(Chromatin Conformation Capture Carbon Copy) method by the ENCODE group
(Dekker Lab) at the University of Massachusetts. The track shows
significant looping interactions between transcriptional start sites
(TSS) and distal regulatory elements in the context of the 44 ENCODE
pilot regions spanning 1% of the human genome.
ENCODE Pilot Regions:
This track depicts the 44 target regions covering 1% of the human genome
defined for the ENCODE pilot project. The hg19 coordinates for these
regions were obtained using liftOver from the hg18 track.
Replication Timing by Repli-chip from ENCODE/FSU:
This track shows genome-wide assessment of DNA replication timing in 7 cell types (3 stem cell, HeLa, IMR90, lymphoblastoid and BG02ES) using NimbleGen tiling CGH microarrays.
Gene Annotations from ENCODE/GENCODE Version 11:
This version features 79% coverage of genes by manual annotation.
It is based on the Ensembl 66 (Feb 2012)
and Vega 46 (Jan 2012) gene predictions and curation.
Transcription Factor Binding Sites by ChIP-seq from ENCODE/HAIB (Release 2):
This release contains 68 new experiments, including 1 new cell line (HUVEC) and 20 new antibodies.
There are also data corrections and new validation datasets (shRNA knockdown).
CTCF Binding Sites by ChIP-seq from ENCODE/University of Washington (Release 3):
This release contains 3 additional experiments in H7ES, NHLF, and HFF cell types.
DNaseI Hypersensitivity by Digital DNaseI from ENCODE/University of Washington (Release 4):
This release contains 11 new experiments across 9 new cell types.
Histone Modifications by ChIP-seq from ENCODE/Broad Institute (Release 2):
This release contains 64 new experiments including 1 new cell line
(Dnd41) and 10 new antibodies, and 3 datasets have additional data.
Open Chromatin by DNaseI HS from ENCODE/Duke (Release 2):
This release provides data in additional cell lines (Colo29, CD4+,
ECC-1, H7ES, HEK293T, 3 IPS lines derived from fibroblasts, MCF-7,
Medullo, RWPE1, T-47D), higher-quality data for the K562 cell line, and
additional data corrections and replacements.
Affymetrix Exon Array from ENCODE/Duke (Release 3):
This track displays human tissue microarray data in 56 cell lines. The
RNA extracted from these cells were also analyzed by DNaseI
hypersensitivity (Duke DNaseI HS track), FAIRE (UNC FAIRE track) and
ChIP-seq (UTA TFBS) by the ENCODE OpenChromatin group.
RNA-seq from ENCODE/Caltech (Release 3):
Signal files for all experiments have been replaced with more accurate data. Earlier versions can still be accessed for download only.
29 Mar 2012 - Mouse ENCODE data releases: PSU TFBS, CSHL Long RNA-seq, LICR Histone (Rel 2), LICR TFBS (Rel 2)
Two new tracks and 2 track updates were released on the mm9 genome browser:
Transcription Factor Binding Sites by ChIP-seq from ENCODE/PSU:
This track shows occupancy of genomic DNA by 5 transcription factors
(CTCF, GATA1, GATA2, PAX5, TAL1) and Pol2 as determined by ChIP-seq in 6
erythroid and other blood cell types (CH12, MEL, G1E, megakaryocyte,
erythroblast, G1E-ER4).
Long RNA-seq from ENCODE/Cold Spring Harbor Lab:
This track contains information about mouse RNAs (> 200 nucleotides in length) in 17 different tissues, as obtained by Illumina squencing.
Histone Modifications by ChIP-seq from ENCODE/LICR (Release 2):
Transcription Factor Binding Sites by ChIP-seq from ENCODE/LICR (Release 2):
These two companion tracks survey cis-regulatory elements in the mouse genome. ChIP-seq was used to localize binding of Pol2, CTCF, and p300 factors and to profile 7 chromatin modification in 20 different mouse (C57Bl/6) tissues, primary cells, and cell lines. Release 2 of these tracks adds 87 new experiments.
24 Feb 2012 - Human ENCODE data releases: UW Repli-seq, HAIB RNA-seq, and 7 track updates
Two new tracks and 7 track updates were released on the hg19 genome browser:
Replication Timing by Repli-seq from ENCODE/University of Washington:
This track shows genome-wide assessment of DNA replication timing in 15
cell lines as identified by the sequencing-based "Repli-seq" method.
Replication timing is known to be an important feature for epigenetic
control of gene expression that usually operates at a higher-order
level than at the level of specific genes.
RNA-seq from ENCODE/HAIB:
This track displays RNA-seq alignments and graphs of signal enrichment
for 9 cell lines, in various treatment protocols. Estimates of
transcript abundance are provided for download.
Gene Annotations from ENCODE/GENCODE Version 10:
This version of the GENCODE Genes track features 77% coverage of
genes by manual annotation. It corresponds to the Ensembl 65 (Dec 2011)
and Vega 45 (Oct 2011) gene predictions and curation.
DNaseI Digital Genomic Footprinting from ENCODE/University of Washington (Release 3):
Release 3 includes experiments in 19 additional cell types.
Histone Modifications by ChIP-seq from ENCODE/University of Washington (Release 3):
This release adds the H7-hESC cell type and another replicate for the GM12865 experiment.
CTCF Binding Sites by ChIP-seq from ENCODE/University of Washington (Release 2):
Release 2 includes experiments in 15 additional cell lines.
DNaseI/FAIRE/ChIP Synthesis from ENCODE/OpenChrom(Duke/UNC/UTA) (Release 2):
Release 2 adds synthesis analysis for 10 cell types:
Gliobla, GM12891, GM12892, GM18507, GM19239, HeLa-S3/IFNa4h, HTR8svn,
Medullo, PanIslets, Urothelia
Small RNA-seq from ENCODE/Cold Spring Harbor Lab (Release 2):
This release adds 10 new cell lines (A549, AG04450, BJ, H1-hESC,
HeLa-S3, HepG2, HUVEC, MCF-7, NHEK, and SK-N-SH_RA), a new displayed
view (Contigs), and two new download files (GENCODE Predicted Exons and
a Protocol Document).
RNA-seq from ENCODE/Caltech (Release 2):
Data was reprocessed with a newer mapping algorithm (labeled V2).
Additional download files of gene, transcript, and exon expression
levels are now provided.
Genotype (CNV and SNP) by Illumina 1MDuo and CBS from ENCODE/HudsonAlpha (Release 2):
This release contains data corrections to GM12878 and astrocyte experiments.
5 Jan 2012 - Human ENCODE data releases: UW Histone, HAIB Methyl450, RIKEN CAGE Loc, UW Affy Exon (Release 2)
Three new tracks and one track update were released on the hg19 genome browser:
Histone Modifications by ChIP-seq from ENCODE/University of Washington:
This track shows genome-wide maps of histone modifications associated with active promoters (H3K4me3), repressed regions (H3K27me3), and active transcription (H3K36me3) in 57 cell types, as identified by ChIP-seq.
CpG Methylation by Methyl 450K Bead Arrays from ENCODE/HAIB:
This track displays the methylation status of specific CpG dinucleotides in 61 cell types as identified by the Infinium Human Methylation 450 Bead Array platform. In general, methylation of CpG sites within a promoter causes silencing of the gene associated with that promoter.
RNA Subcellular CAGE Localization from ENCODE/RIKEN:
This track from the ENCODE Transcriptome group shows 5' cap analysis gene expression (CAGE) tags and clusters.
A total of 34 Experiments were conducted in 12 cell lines and one tissue (prostate), with RNA extracted from 6 isolated cellular compartments and in whole cell.
Affymetrix Exon Array from ENCODE/University of Washington (Release 2):
This track from the ENCODE Transcriptome group shows human tissue miroarray data from the Affymetrix Human Exon 1.0 GeneChip.
Release 2 of this track shows experiments in 14 additional cell lines.
1 Dec 2011 - Mouse ENCODE data releases: Caltech TFBS, Caltech Histone
Two new tracks were released on the mm9 genome browser:
Transcription Factor Binding Sites by ChIP-seq from ENCODE/Caltech:
This track shows 15 ChIP-seq experiments mapping 7 histone marks in the C2C12 myoblast cell line; both untreated cells and cells under timecourse treament with insulin to stimulate myogenesis.
Histone Modifications by ChIP-seq by ChIP-seq from ENCODE/Caltech:
This track shows 26 ChIP-seq experiments mapping 14 transcription factors in the C2C12 myoblast cell line; both untreated cells and cells under timecourse treament with insulin to stimulate myogenesis.
10 Nov 2011 - ENCODE portal changes: New File Formats page and updates to Publications, Data Summaries, and Data Standards
The ENCODE portal was updated to include informative new and expanded pages.
ENCODE-related data file formats are now documented on the new
File Formats page.
The
Publications page was expanded to a comprehensive list of Consortium
publications, plus methods, resource, and biological findings papers by
ENCODE-funded projects.
The
Human Data Summary and
Mouse Data Summary spreadsheets detailing ENCODE submissions were updated to reflect
status as of the latest ENCODE quarterly reporting (Sept. 30 2011).
Finally, the
Data Standards section was greatly expanded to include new standards documents (v2.0 ChIP-seq) and a new
Platform Characterization section.
29 Oct 2011 - Human ENCODE data releases: SYDH TFBS, CSHL Long RNA-seq, GIS RNA
PET, UW DNaseI HS (release 3), Mappability (Release 3)
Three new tracks and two track updates were released on the hg19 genome browser:
Transcription Factor Binding Sites by ChIP-seq from Stanford/Yale/USC/Harvard:
This comprehensive track shows 306 ChIP-seq experiments identifying transcription factor binding sites in
19 cell types.
Long RNA-seq from ENCODE/Cold Spring Harbor Lab:
This comprehensive track from the ENCODE Transcriptome group shows RNAs longer than 200 nucleotides. Profiling was performed on RNA extracts enriched and depleted for polyA+, in multiple cellular compartments and whole cell, in 15 cell lines.
RNA Subcellular Localization by Paired End diTag Sequencing from GIS/ENCODE:
This track from the ENCODE Transcriptome group shows starts and ends of full length mRNA transcripts determined by GIS paired-end ditag (PET) sequencing.
Experiments were conducted in 7 cell lines and one tissue (prostate), with RNA extracted from 6 isolated cellular compartments and in whole cell.
19 Sept 2011 - Mouse ENCODE data releases: Stan/Yale Histone, LICR TFBS, LICR RNA-seq, Stan/Yale TFBS (Release 2)
Three new tracks and one track update were released on the mm9 genome browser:
Histone Modifications by ChIP-seq from ENCODE/Stanford/Yale:
This track shows locations of histone H3K4me3 modification in CH12 and MEL cell types.
The two cell lines were selected for similarity to the ENCODE Tier1 cell lines (GM12878 and K562).
Transcription Factor Binding Sites by ChIP-seq from ENCODE/LICR, and
RNA-seq from ENCODE/LICR:
These tracks show transcriptome and Pol2, p300 and CTCF binding sites in mouse bone marrow, cerebellum, cortex, heart, kidney, liver, lung, spleen, embryonic fibroblast cells (MEFs) and embryonic stem (ES) cells.
Transcription Factor Binding Sites by ChIP-seq from ENCODE/Stanford/Yale (Release 2):
This release adds binding site localization for 13 additional transcription factors to the previously released TFBS track.
15 Sept 2011 - Summer ENCODE human data releases: GENCODE Genes V7, HAIB TFBS, Stanf Nucleosome, HAIB Genotype, SUNY RIP-seq, SUNY Switchgear
Six tracks of human ENCODE data were released in late summer on the hg19 genome browser:
Gene Annotations from ENCODE/GENCODE Version 7:
The GENCODE Version 7 Genes track shows high-quality manual annotations merged with evidence-based automated annotations across the entire human genome. This version of GENCODE provides an increase of 25% in manual curation of transcripts over the previous (V4) version at UCSC.
Transcription Factor Binding Sites by ChIP-seq from ENCODE/HudsonAlpha:
This track displays 185 experiments identifying transcription factor binding sites in multiple cell lines by chromatin immunoprecipitation followed by high throughput sequencing. A total of 56 transcription factor target antibodies and 20 cell types are represented.
Nucleosome Position by MNase-seq from ENCODE/Stanford/BYU:
This track displays nucleosome position density maps from micrococcal nuclease digested chromatin in GM12878 and K562 cell lines.
In the context of the ENCODE project, nucleosome positioning data are particularly valuable for analysis of the relationship between transcription factor binding, histone modifications, and gene activity.
Genotype (CNV and SNP) by Illumina 1MDuo and CBS from ENCODE/HudsonAlpha:
This track displays copy number variation (CNV) as determined by the Illumina Human 1M-Duo Infinium HD BeadChip assay and circular binary segmentation (CBS). Allele frequency and single nucleotide polymorphism (SNP) data generated by the experiment are available for download.
RIP-seq from ENCODE/SUNY Albany:
This track displays transcriptional fragments associated with RNA binding proteins in K562 and GM12878 cell lines, using Ribonomic profiling followed by high throughput sequencing.
RNA Binding Protein Associated RNA by SwitchGear from ENCODE/SUNY Albany:
This track displays 3' UTR regions associated with RNA binding proteins in the HT-1080 cell line as evidenced by reporter assays.
8 July 2011 - Mouse ENCODE data releases: DNaseI hypersensitivity (UW DNaseI HS) and histone modifications (LICR Histone)
Two tracks of ENCODE data were released on the mm9 genome browser, from the UCSD/Ludwig Institute for Cancer Research and the University of Washington Mouse ENCODE groups.
Histone Modifications by ChIP-seq from ENCODE/LICR:
This track shows a comprehensive survey of cis-regulatory elements in the mouse genome by using ChIP-seq to identify transcription factor binding sites and chromatin modification profiles in many mouse tissues and primary cells, including bone marrow, cerebellum, cortex, heart, kidney, liver, lung, spleen, mouse embryonic fibroblast cells (MEFs) and embryonic stem (ES) cells.
DNaseI Hypersensitivity by Digital DNaseI from ENCODE/University of Washington:
This track shows DNaseI sensitivity measured genome-wide in mouse tissues and cell lines using the Digital DNaseI methodology and DNaseI hypersensitive sites.
1 July 2011 - ENCODE data releases: Broad ChromHMM, Open Chrom Synth, UChicago TFBS, Duke Affy Exon
Four tracks of ENCODE production data and analysis were released in June, from the
Broad Institute (Kellis lab), OpenChromatin (Duke, UNC, UT-A) and University of Chicago
(White Lab) ENCODE groups. This is the first data release from the University of Chicago ENCODE group,
which joined the Consortium as part of the NIH ARRA stimulus grants.
Chromatin State Segmentation by HMM from ENCODE/Broad:
This track, and the companion hg18 track, display chromatin state segmentation of the human genome into
fifteen states grouped to predict functional elements.
DNaseI/FAIRE/ChIP Synthesis from ENCODE/OpenChrom(Duke/UNC/UTA):
This track displays a synthesis of open chromatin regions and binding of selected regulatory factors,
based on three complementary methodologies.
Transcription Factor Binding Sites by Epitope-Tag ChIP-seq from ENCODE/University of Chicago:
This track maps human transcription factor binding sites genome-wide using expressed transcription factors as GFP tagged fusion proteins after BAC recombineering.
Affymetrix Exon Array from ENCODE/Duke:
This track displays human tissue microarray data using Affymetrix Human Exon 1.0 ST expression arrays,
including annotations at the gene level.
24 June 2011 - ENCODE RNA-seq data standards now available
The ENCODE Consortium has finalized 'Standards, Guidelines and Best Practices for RNA-Seq V1.0', as part of the Consortium's continuing
effort to generate data standards. The document is available at the ENCODE portal via the
Data Standards link.
RNA-Seq is a directed experimental approach aimed at characterizing transcription in biological samples. This document presents a set of guidelines and standards focused on best practices for creating 'reference quality' transcriptome measurements.
sets.
1 June 2011 - ENCODE data releases in April and May
Five tracks of ENCODE production and analysis data were released in April and May on the GRCh37/hg19 human assembly from
the Caltech, Broad Institute, HudsonAlpha Institute for Biotechnology, Duke University (Open Chromatin), SUNY Albany,
University of Washington, Boston University, Stanford/Yale/Davis/Harvard and UCSC ENCODE groups
Integrated Regulation from ENCODE:
This collection of tracks displays integrated signal and clustering annotations from multiple cell lines, using ENCODE
primary data from RNA-seq, ChIP-seq, and DNase-seq assays. This track is a companion to the hg18 ENCODE Regulation track.
Open Chromatin by DNaseI HS from ENCODE/OpenChrom(Duke University):
This track displays DNaseI hypersensitivity evidence as part of the four Open Chromatin track set.
RNA Binding Protein Associated RNA by RIP-chip GeneST from ENCODE/SUNY Albany:
This track displays transcriptional fragments associated with RNA
binding proteins in different cell lines using RIP-Chip (Ribonomic)
profiling on Affymetrix GeneChip® Human Gene 1.0 ST Arrays.
DNaseI Hypersensitivity by Digital DNaseI form ENCODE/University of Washington:
This track shows DNaseI sensitivity measured genome-wide in different
cell lines using the Digital DNaseI methodology.
OrChID Predicted DNA Cleavage Sites from ENCODE/Boston Univ (Tullius Lab):
This track display predicted hydroxyl radical cleavage intensity on naked DNA for each nucleotide in the genome.
Histone Modifications by ChIP-seq from ENCODE/Stanford/Yale/Davis/Harvard:
This track displays maps of histone modifications genome-wide using ChIP-seq in different cell lines.
22 April 2011 - ENCODE data releases: UTA TFBS, UW CTCF, UNC FAIRE, RIKEN CAGE
Loc, UW Affy Exon, UW DNaseI DGF & Duke DNaseI HS
Seven tracks of ENCODE data on the GRCh37/hg19 human assembly were released in March and April from
the University of Texas at Austin (Open Chromatin), University of Washington, University of North Carolina (Open
Chromatin), RIKEN, and Duke University (Open Chromatin) ENCODE groups (6 of the tracks are new to hg19, and one is a Release 2 on hg19):
UTA TFBS:
This track displays chromatin immunoprecipitation (ChIP-seq) evidence as part of the four Open
Chromatin track sets.
UW CTCF:
This track displays maps of genome-wide binding of the CTCF transcription factor in different cell
lines using ChIP-seq high-throughput sequencing.
UNC FAIRE:
This track displays Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE) evidence as part
of the four Open Chromatin track sets.
RIKEN CAGE Loc:
This track shows 5' cap analysis gene expression (CAGE) tags and clusters in RNA extracts from
different sub-cellular localizations in multiple cell lines.
UW Affy Exon:
This track displays human tissue microarray data using Affymetrix Human Exon 1.0 GeneChip.
UW DNaseI DGF (Release 2):
This track contains deep sequencing DNase data that will be used to identify sites where regulatory
factors bind to the genome (footprints).
Duke DNaseI HS:
This track displays DNaseI hypersensitivity (HS) evidence as part of the four Open Chromatin track
sets.
14 February 2011 - ENCODE data releases: Caltech RNA-seq, Broad Histone, UW Histone & SUNY RIP Tiling
Four tracks of ENCODE data on the GRCh37/hg19 human assembly
from the Caltech, Broad/MGH, SUNY Albany and University of Washington ENCODE groups were released:
Caltech RNA-seq:
This track shows transcriptome measurements performed on polyA+ RNA using both stranded and unstranded protocols.
Broad Histone,
UW Histone:
These tracks display maps of histone modifications reflective of chromatin state changes identified by ChIP-seq.
SUNY RIP Tiling:
This track displays transcriptional fragments associated with RNA binding proteins in different cell lines,
using RIP-Chip (Ribonomic) profiling on Affymetrix GeneChip ENCODE 2.0R Tiling Arrays.
9 February 2011 - First Mouse ENCODE data release: Transcription Factor Binding Sites by ChIP-seq from Stanford/Yale
The first Mouse ENCODE data is now available on the mm9 (NCBI37) genome assembly. The
Stan/Yale TFBS track
in the 'Regulation' track group shows probable binding sites of the following transcription
factors: c-MYB (H-141), CTCF (C-20), Max, NELFE, p300 (N-15), Rad21, and
USF2, in the MEL leukemia (K562 analog) cell line as determined by ChIP-seq.
Thanks to all who had a hand in generating this data and to the UCSC
wrangler, Venkat Malladi, and Q/A staff who made this data release
possible.
16 December 2010 - Release of DNA Methylation and DNaseI Sensitivity data
Three tracks of ENCODE DNA Methylation and DNaseI Sensitivity data have been released on the GRCh37/hg19 human
assembly. All three tracks are in the browser 'Regulation' track group; one of the tracks is in the
new
ENC DNA Methyl super-track and the other two are in the new
ENC DNase/FAIRE
super-track (super-tracks provide additional documentation and organization by data type). The three
newly released tracks are:
HAIB Methyl
RRBS: This track reports the percentage of DNA molecules that exhibit cytosine methylation
at specific CpG dinucleotides. In general, DNA methylation within a gene's promoter is associated
with gene silencing, and DNA methylation within the exons and introns of a gene is associated with
gene expression.
UW DNaseI DGF:
This track contains deep sequencing DNase data that will be used to identify sites where regulatory
factors bind to the genome (footprints).
UW DNaseI HS:
This track shows DNaseI sensitivity measured genome-wide in different cell lines using the Digital
DNaseI methodology and DNaseI hypersensitive sites.
Some of the data that comprise these tracks were originally released on hg18 and have been remapped
to hg19; in such cases, subtracks have 'origAssembly hg18' as part of their metadata.
16 November 2010 - Release of the first ENCODE RNA-seq data on hg19
We are pleased to announce the release of the first ENCODE RNA-seq data on the GRCh37/hg19 human
browser. Two tracks have just been released; these are organized in the new
ENC RNA-seq super-track
within the browser 'Expression' track group. The super-track provides additional documentation and
organization by data type. The two tracks released are:
CSHL Sm
RNA-seq: This track depicts NextGen sequencing information for RNAs between the sizes of
20-200 nt isolated from RNA samples from tissues or subcellular compartments from ENCODE cell lines.
GIS RNA-seq:
This track shows high throughput sequencing of RNA samples from tissues or subcellular compartments
from cell lines included in the ENCODE Transcriptome subproject.
All of the data that comprise these tracks were originally released on hg18 and have been remapped
to hg19.
15 Nov 2010 - New ENCODE Tutorial at OpenHelix
OpenHelix, together with the UCSC Genome Bioinformatics group, anounce a new online
tutorial suite to teach users how to access the ENCODE data in the UCSC Genome
Browser. This tutorial introduces the types of data available under ENCODE, and
presents methods to access the data via the Genome Browser, Table Browser, and
downloads. This tutorial suite is freely available at
OpenHelix
16 September 2010 - First Production ENCODE Data on hg19 has been Released
We are pleased to announce the release of the first sets of production ENCODE data
on hg19:
GIS DNA PET: This track shows the starts and ends of DNA fragments from
different cell lines determined by paired-end ditag (PET) sequencing using different
DNA fragment sizes for analysis of genome structural variation. The data in this
track uses the new BAM data format. For more information about SAM/BAM, click
here. All of the subtracks
that comprise this track were originally released on hg18 and have been remapped to
hg19.
Gencode Genes:
This track (version 4, May 2010) shows high-quality manual annotations merged with
evidence-based automated annotations across the entire human genome generated by the
GENCODE project. Previous versions of this data were released on hg18, but this
newest version is available solely on hg19.
20 August 2010 - New ENCODE Integrated Regulation Super-track Released
We are pleased to announce the release of the ENCODE Integrated
Regulation super-track, a collection of regulatory tracks containing
state-of-the-art information about the mechanisms that turn genes on and
off at the transcription level. Individual tracks within the set show
enrichment of histone modifications suggestive of enhancer and promoter
activity, DNAse clusters indicating open chromatin, regions of
transcription factor binding, and transcription levels. When viewed in
combination, the complementary nature of the data within these tracks
has the potential to greatly facilitate our understanding of regulatory
DNA.
The data comprising these tracks were generated from hundreds of
experiments on multiple cell lines conducted by labs participating in
the ENCODE project, and were submitted to
the UCSC ENCODE Data Coordination Center for display on the Genome
Browser.
Faced with the problem of how to display such a large amount of data
in a manner facilitating analysis, UCSC has developed new visualization
methods that cluster and overlay the data, and then display the
resulting tracks on a single screen. Each of the cell lines in a track
is associated with a particular color. Light, saturated colors are used
to produce the best transparent overlay.
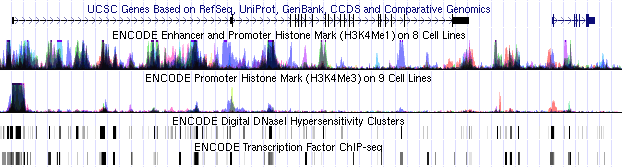
Currently, the ENCODE Regulation data are available only on the
March 2006 (NCBI Build 36, UCSC version hg18) assembly of the human
genome.
For a detailed description of the datasets contained in this super-track
and a discussion of how the tracks can be used synergistically to
examine regions of regulatory functionality within the genome, see the
track description page.
6 August 2010 - June and July ENCODE news
Initial Release of the
HudsonAlpha RNA-seq
track: This track shows short
tag sequencing of cDNA obtained from biological replicate samples
(different culture plates) of the ENCODE cell lines. The sequences
were aligned to the human genome (hg18) and UCSC known-gene splice
junctions.
Release 2 of the
Caltech RNA-seq
track: This track shows alignments, signal density, and
splice sites based on 75 bp paired reads and 32 bp strand-specific
single reads of polyA+ RNA aligned to the human genome (hg18) and
UCSC known-gene splice junctions. Also included with the track as
downloadable files are RPKM expression level measurements at the
gene-level and exon-level, and candidate novel exons. Release 2
of this track adds five new cell types: H1-hESC, HeLa-S3, HepG2,
HUVEC, and NHEK.
Initial Release of the
GIS PET Loc
track: This track shows starts and ends of full length
mRNA transcripts determined by PET sequencing of polyA+ and total
RNA from 6 subcellular compartments and whole cell, in 8 cell lines.
Release 2 of the HAIB TFBS
track: This track shows signal density and binding sites
of selected transcription factors in a variety of cell types.
Release 2 of this track adds 73 new experiments covering 13 new cell
lines and 27 antibodies. Additionally, DEX and EtOH treatments have
been included in the A549 cell line.
Initial Release of the BU ORChID
track: This track displays the predicted hydroxyl
radical cleavage intensity on naked DNA for each nucleotide in the
genome.
Update of the Mappability
track: This track displays the level of sequence
uniqueness of the reference hg18 genome. The update adds CRG
Alignability data, which displays how uniquely k-mer sequences align
to a region of the genome.
19 July 2010 - April and May ENCODE news
Initial Release of the GIS RNA-seq
track: This track shows RNA-seq of high-quality PolyA+
RNA in ENCODE Tier1 cell lines and H1 ESC sequenced on the ABI Solid
platform.
Release 3 of the Yale TFBS
track: This track shows probable binding sites of the
specified transcription factors (TFs) in the given cell types as
determined by chromatin immunoprecipitation followed by high
throughput sequencing (ChIP-Seq). Release 3 adds 64 experiments
and 26 input/control datasets for a total of 54 factors in 21 cell lines.
Release 2 of the RIKEN CAGE Loc
track: This track shows 5' cap analysis gene expression
(CAGE) tags and clusters in RNA extracts from different sub-cellular
localizations. Release 2 of this track adds data for eight new
cell-type/compartment combinations (GM12878 Nucleus, H1-hESC whole
cell, HepG2 cytosol/nucleus/nucleolus, HUVEC cytosol, and NHEK
cytosol/nucleus).
Initial Release of the Duke Affy Exon
track: This track shows gene expression by microarray of
RNA extracted from 28 cell lines that were also analyzed by DNaseI
hypersensitivity, FAIRE, and ChIP assays (Open Chromatin track).
Initial Release of the UW DNase DGF
track: This track shows high-resolution DNase
annotations from samples sequenced to depths of 300-fold or greater,
in 5 cell lines.
Initial Release of the GIS DNA PET
track: This track shows the starts and ends of DNA
fragments from different cell lines determined by paired-end ditag
(PET) sequencing using different DNA fragment sizes for analysis of
genome structural variation.
18 March 2010 - February and March 2010 ENCODE news
Release 3 of the Open Chromatin
track: This track displays evidence of open chromatin in
multiple cell types from the Duke/UNC/UT-Austin/EBI ENCODE group.
Release 3 of this track includes 18 new cell line or cell/treatment
experiments. In addition, a number of new experiments were added to
existing cell lines. Almost all Peaks have been called anew using
improved cut-offs and p-Values. Finally, a second type of peak
called using a ZINBA algorithm has been provided for several of the
FAIRE-seq experiments.
Release 3 of the Broad Histone
track: This track shows maps of chromatin state
generated using CHIP-seq. Release 3 of this track adds the HSMM cell
line and includes new experiments for H1-hESC and NHLF.
Release 2
of the UW Affy Exon track:
This track displays
human tissue microarray data using the
Affymetrix Human Exon 1.0 GeneChip. This release includes 28 new cell types, and replaces the data for four existing tables (replicate 1 for K562, NB4, and SKMC; replicate 2 for HeLa-S3).
Initial release
of the UW Histone track:
This track displays
maps of histone modifications genome-wide in different cell lines, using ChIP-seq high-throughput sequencing.
Release 2 of the HudsonAlpha
Methyl-seq track:
Release 2 adds data for five new cell
types.
Release 3 of the Gencode
Genes track: shows high-quality
manual annotations in the ENCODE regions generated by the
GENCODE
project.
Version 3 of the
Gencode gene set presents a full merge between HAVANA and ENSEMBL,
giving priority to the manually curated Havana objects and using
ENSEMBL objects where they are different or fall into un-annotated
regions.
Initial release of the CSHL
Small
RNA-seq track: This track depicts
NextGen sequencing information for RNAs between the sizes of 20-200 nt
isolated from RNA samples from tissues of sub cellular compartments
from ENCODE cell lines.
Release 3 of the UW
DNaseI HS track:
This track shows
DNaseI sensitivity measured genome-wide in different using the Digital DNaseI
methodology, and DNaseI hypersensitive sites. This
release includes 19 new cell lines as
well as new version of NB4 replicate 1.
6 January 2010 - December 2009 ENCODE news
"ENCODE whole-genome data in the UCSC Genome Browser":
This paper addresses the history of the ENCODE project, summarizes the datasets
available as of September 2009, and outlines methods to access the data. See
Nucleic Acids Res. 2010 Jan;38(Database issue):D620-5.
Initial release of the
Caltech RNA-seq
track: This track contains sequence reads and RPKM transcript abundance
measures for sequences that map to either the genome or to known RNA splice sites.
The results of four different mapping algorithms are provided, enabling
comparison between different mapping algorithms. Results are available for
polyA+ and total RNA for the two ENCODE Tier 1 cell lines.
Release 2 of the
Broad Histone
track: This track displays maps of chromatin state generated using CHIP-seq.
Release 2 adds data for the ENCODE Tier 2 cell lines H1-hESC and HepG2, plus
NHLF (normal human lung fibroblasts) and HMEC (human mammary epithelial) cells. This
expands the track data to 9 cell lines, and 11 antibodies plus an input control.
Release 2 of the
CSHL Long RNA-seq
track: This track depicts sequencing of long RNAs of more than 200
nucleotides in length. Release 2 adds data from strand-specific assays of
total RNA for the two ENCODE Tier 1 cell lines.
Release 2 of the
ENCODE Open Chromatin
track: This track displays evidence of open chromatin as identified by two
complementary methods, DNaseI hypersensitivity and FAIRE, combined with ChIP
identification methods. Release 2 adds data from eight additional cell types,
expanding the track to 41 experiments in 13 cell lines.
7 November 2009 - October ENCODE News
Sep 2009 data freeze complete: The ENCODE Consortium has just
completed data submissions for the fourth production data freeze (Sep 09). The first
set of data from this freeze to complete quality review is now available on the UCSC
public server, in Release 2 of the
ENCODE Transcription Factor Binding Sites from Yale/UC-Davis/Harvard
track. Release 2 adds 59 ChIP-seq experiments to this track.
Other October track releases: The
Affymetrix/CSHL Subcellular RNA Localization by Tiling Array
track was expanded to include 4 additional experiments.
encodeproject.org: By request of the ENCODE Consortium, the domain
encodeproject.org has been registered by the ENCODE Data Coordination Center,
and is redirected to the ENCODE portal at UCSC.
New grants funded: NHGRI has funded 5 new ENCODE grants, as part
of the American Investment and Recovery Act. The new grants include expansion of
ENCODE to the mouse genome and proteogenomics.
Job openings at UCSC: The UCSC Genome Browser and ENCODE projects
are currently accepting applications for
Software Developer and
Biological Database Testing/User Support Technician
positions. We are looking for talented individuals who would like to use their
skills in computer science, biology, and bioinformatics on fast-paced projects
featuring the work of top genomics scientists worldwide.
24 September 2009 - ENCODE data releases since July 1
During this period a total of 10 new ENCODE tracks were released to the UCSC public
server. Functional elements and region characterization in these tracks include:
For track names and file access, see the Release Log and Downloads
links listed in the left menu bar.
We would like to thank the contributing ENCODE labs and the the DCC team at UCSC for their efforts
completing these tracks.
1 July 2009 - ENCODE data releases for the period April - June 2009
The following ENCODE tracks were released to the ENCODE DCC/UCSC public server during this period:
The Release Log listed in the left menu bar shows all released ENCODE tracks.
We would like to thank the HudsonAlpha, Transcriptome, Open Chromatin, Broad, and SUNY ENCODE
groups and the DCC team at UCSC for their efforts completing these tracks.
13 March 2009 - ENCODE Data Release: Transcription Factor Binding Sites from Yale/UC-Davis/Harvard
We are pleased to announce the release to the ENCODE DCC/UCSC public server
of the
ENCODE Transcription Factor Binding Sites by ChIP-seq from Yale/UC-Davis/Harvard
(Yale TFBS, in the Regulation group).
This track shows probable binding sites of 12 transcription factors
and RNA polymerase II in 7 cell types, as determined by chromatin
immunoprecipitation followed by high-throughput sequencing.
The Genome Browser displays discrete Peaks
of enrichment and Signal graphs of enrichment density for these
experiments.
The sequence reads, quality scores, and sequence alignment coordinates from
these experiments are available for
download.
We would like to acknowledge the efforts of the Yale/UC-Davis/Harvard
ENCODE group and the work of the UCSC data wrangler for this group,
Tim Dreszer, for completing this track. We also thank
the entire UCSC ENCODE team and the UCSC Quality Assurance
group for contributing to this first ENCODE track release.
27 Feb 2009 - ENCODE February 2009 data freeze
The February 2009 ENCODE data freeze supplements the data contributed for the
November 2008 freeze, and will be used together with the earlier freeze
for the initial analysis effort of the ENCODE Consortium. Data from
this freeze is being incorporated into tracks created from the first
data freeze, and is being reviewed by the UCSC Quality Assurance team.
9 Dec. 2008 - First ENCODE whole-genome data freeze completed
The ENCODE Consortium has just completed the first freeze
(November 2008) of whole-genome experimental data
produced for the ENCODE production phase. Data submitted
to the DCC for this freeze include:
- transcription factor binding sites
- histone modifications
- DNaseI hypersensitive sites
- DNA methylation
- transcription maps and tags, localized to subcellular
compartments
- GENCODE gene annotations
Experiments during this freeze focused on the ENCODE
Tier1 cell lines -- K562 leukemia, and GM12878 lymphoblastoid
(which is also a
1000 Genomes project
sample designated for in-depth analysis of genetic variation).
The freeze also includes data
from some ENCODE Tier2 and Tier3 cell lines (see
Cell Types). The majority
of these experiments were assayed by high-throughput
sequencing (ChIP-seq, DNase-seq, and RNA-seq).
The UCSC quality team is currently reviewing these data.
When the review is complete, the browser tracks and
associated downloads will be released to the UCSC public
Genome Browser.
Thanks to the many labs who contributed data for the
initial phase of this project. We'd also like to acknowledge
the UCSC ENCODE team for data wrangling during the freeze,
and for the development and maintenance of
the ENCODE automated data submission pipeline and
associated tools: Kate Rosenbloom, Tim Dreszer, Larry Meyer,
Michael Pheasant, Ting Wang, Galt Barber, and Andy Pohl.
4 Oct. 2007 - ENCODE Genome Browser Released for hg18 Assembly
The ENCODE browser for UCSC human genome assembly hg18
(NCBI Build 36) is now available. You can access the browser
directly at
http://genome.ucsc.edu/ENCODE/encode.hg18.html
or by clicking the ENCODE link on the Genome Browser
home page, then
selecting the Regions (hg18) item in the sidebar menu on
the ENCODE portal page.
The hg18 ENCODE browser includes 540 data tables in 59
browser tracks that were migrated from the hg17 browser.
The hg17 data coordinates were converted to hg18 coordinates
using the UCSC liftOver process.
To improve the accessibility of the data, related ENCODE
tracks have been gathered into new configuration groupings
("super-tracks") that can be displayed or hidden
using a single visiblity control. We have also reduced the
number of track groups and have modified some of the group
names for clarity.
The following table
summarizes the data currently present in the hg18 ENCODE
browser:
| Group |
Super-tracks |
Tracks |
Tables |
| Regions and Genes | 2 | 12 | 73 |
| Transcription | 2 | 11 | 67 |
| Chromatin Immunoprecipitation | 8 | 28 | 349 |
| Chromatin Structure | 2 | 8 | 51 |
Note that the Variation and Comparative Genomics data were
not lifted during this migration; instead, they will be
replaced by new data. The first ENCODE MSA alignment for
hg18 (TBA) is currently in progress on the UCSC
development
server.
During the migration, ENCODE tracks with whole-genome
data were moved into the standard browser track
groups. These include the GIS PET and UCSD/LI
TAF1 tracks. Future submissions of whole-genome ENCODE data
will be loaded directly into the standard track groups.
We have expanded the ENCODE downloads site to include
original data for all "wiggle" datasets. These
data files now have filename extensions indicating the
wiggle input format (fixed step, variable step, or
bedGraph).
You can find a description of the migration project and
full details of the tables, tracks, and super-tracks
available at the UCSC ENCODE portal on the UCSC
genomeWiki.
The UCSC team members who contributed to this effort were:
Andy Pohl (data conversion and database loading), Ting Wang
and Donna Karolchik (super-track documentation), Bob Kuhn
(portal updates), Brooke Rhead, Kayla Smith, and Ann Zweig (quality
assurance), and Kate Rosenbloom (super-track development,
project management).
13 Jun. 2007 - ENCODE Findings Published in Nature and Genome Research
The findings of the ENCODE project have been
released to the public today, the culmination of a
four-year effort to catalog the biologically functional
elements in 1 percent of the human genome. The publications,
which include a group paper in the 14 June 2007 issue of
Nature and 28 companion papers
in the June 2007 issue of
Genome Research, were authored by
researchers from academic, governmental, and industry
organizations located in 11 countries. The Nature
issue includes a pull-out poster featuring a screenshot of
the UCSC Genome Browser displaying a broad range of the
ENCODE data.
In the press release accompanying the publication
rollout,
NHGRI Director Francis S. Collins is quoted as saying
"This impressive effort has uncovered many exciting
surprises and blazed the way for future efforts to explore
the functional landscape of the entire human genome. Because
of the hard work and keen insights of the ENCODE consortium,
the scientific community will need to rethink some long-held
views about what genes are and what they do, as well as how
the genome's functional elements have evolved. This could
have significant implications for efforts to identify the
DNA sequences involved in many human diseases."
For more information on the ENCODE project,
including the consortium's data release and accessibility
policies and a list of NHGRI-funded participants, see the
NHGRI ENCODE website.
12 Jun. 2007 - Spring 2007 ENCODE News
Between January and May of 2007, several new or upgraded
data tracks were released by UCSC:
- Gencode March 2007 Genes and Gencode
RACEfrags --
Reannotation of 69 loci consisting of 132 transcripts
based on RACE, array, and sequencing analyses. New features
include the addition of PolyA features, polymorphic
gene type, and integration of experimental intron
validation.
- Yale ChIP-chip RFBR --
Analysis of ChIP-chip data to identify regulatory factor
binding regions, resulting from over 105 experiments
representing 29 transcription factors in 9 cells lines,
performed in 7 labs using 3 microarray platforms. Data
provided by the ENCODE Transcriptional Regulation analysis
group.
- University of Virginia DNA Replication Origins --
Three new datasets representing two new experimental
methods for origin mapping (bubble trapping and nascent
strand) in HeLa and GM06990 cells.
- University of Vienna RNAz --
RNA secondary structure prediction as predicted by RNAz,
based on evolutionary conservation and thermodynamic
stability.
- Duke DNaseI Hypersensitivity --
DNase-chip results in IMR90 (fibroblast), K562 (leukemia)
and H9 (stem) cells.
- UCSC EvoFold -- Replaces TBA23 EvoFold.
- UC Davis ChIP Hits -- Updated cMyc and E2F1.
Thanks to the UCSC staff who worked on these tracks:
Rachel Harte and Kate Rosenbloom (development), and Ann
Zweig, Archana Thakkapallayil, and Kayla Smith (quality
review).
9 Jan. 2007 - Winter 2006 ENCODE News
To improve communication, we have posted instructions for
our ENCODE ftp site on the ENCODE Wiki and have set up an
email alias for notifying UCSC about your ENCODE data
submissions: encode@soe.ucsc.edu.
The current UCSC recipients are Kate Rosenbloom, Daryl
Thomas, Ting Wang, and Rachel Harte.
During November and December, four new/improved data tracks
were released:
-
Chip-PET from the Genome Institute of Singapore -
Genome-wide data for c-Myc in P493 B cells was added as a
new subtrack (cMyc P493, encodeGisChipPetMycP493) to the
existing GIS ChIP-PET track.
-
DNaseI Hs from Duke University -
Existing NHGRI data and new data from the Crawford lab at
Duke University (raw and p-value data for the HepG2 cell
line) were merged into the Duke/NHGRI DNase track. The
newer data is based on DNase-chip technology.
-
STAGE tags from University of Texas -
Raw tags data for STAT1 in HeLa cells were added
as a new subtrack (UT STAT1 HeLa Tags,
encodeUtexStageStat1HelaTags)
to the existing UT-Austin Stage track.
-
DNaseI Hs from University of Washington -
The existing three UW/Regulome DNaseI Sens tracks were
replaced with a single new track (UW/Reg QCP DNaseI Sens)
based on quantitative chromatin profiling (QCP) methods in
16 cell types.
Thanks to the UCSC staff who worked on these tracks:
Rachel Harte, Kate Rosenbloom, and Hiram Clawson
(development), Ann Zweig, Brooke Rhead, and Kayla Smith
(quality review).
7 Oct. 2006 - Comparative Genomics Data Release
Twelve tracks of data produced by the ENCODE Multi-Species
Sequence Analysis group have been released to the UCSC
public server. These tracks contain multiple sequence
alignments, conservation, and conserved (constrained)
elements produced by four conservation methods
(phastCons, binCons, GERP, SCONE) applied to three sequence
alignments (TBA, MLAGAN, MAVID), and also an assessment of
the agreement among the alignment methods. The alignments
were based on genomic sequence in the ENCODE regions of 28
vertebrate species, as defined in the
MSA September 2005 sequence freeze.
The following tracks can now be found in the ENCODE
Comparative Genomics track group on the public ENCODE
browser:
- MSA Consensus Constrained Elements
- TBA Alignments, TBA Conservation, TBA Conserved Elements
- MLAGAN Alignments, MLAGAN Conservation, MLAGAN Conserved
Elements
- MAVID Alignments, Conservation, Conserved Elements
- MSA Alignment Agreement, MSA Alignment Gaps
Thanks to the following providers of this data:
- Elliott Margulies (NHGRI) - TBA alignments, binCons and
phastCons conservation, Consensus elements, sequence freeze
- Daryl Thomas (Haussler lab, UCSC) - sequence freeze
- George Asimenos (Batzoglou lab, Stanford University) -
MLAGAN alignments
- Colin Dewey (while at the Pachter lab, UC Berkeley) -
MAVID alignments
- Greg Cooper (while at the Sidow lab, Stanford
University) - GERP conservation
- Saurabh Asthana (Sunyaev lab, Harvard Genetics) -
SCONE conservation
- Ariel Schwartz (Pachter lab, UC Berkeley) - Alignment
agreement
Also, thanks to the UCSC team that produced these tracks in
the browser: Kate Rosenbloom (track development), Ann Zweig,
Kayla Smith, and Archana Thakkapallayil (quality review).
31 Aug. 2006 - Summer ENCODE data activity
Since mid-June, UCSC has released new ENCODE data from three
labs (Sanger Institute, Uppsala University, and University
of North Carolina) and has a track in
progress for newly submitted data from a fourth lab
(NHGRI/Duke University):
-
Sanger ChIP-chip (MOLT4 and PTR8 cells) - Eight new
datasets were added to the Sanger Chip/chip track.
The new data show sites of H3 histone methylation and
acetylation in MOLT4 (lymphoblastic leukemia) and also
in the chimpanzee PTR8 cell line, used for comparative
analysis. Thanks to Rob Andrews at the Ian Dunham lab
for providing these data.
-
Uppsala University Chip/chip Butyrate - This
track shows the effects of Na-butyrate treatment of
HepG2 (liver carcinoma) cells on histone H3 and H4
acetylation, assayed on Sanger microarrays. Thanks to Adam
Ameur at the Claes Wadelius lab for providing these data.
-
University of North Carolina FAIRE (Peaks data
update) -
This track was updated to include a subtrack of peaks
generated by an alternate peak-finding algorithm,
ChIPOTle. The FAIRE data were generated from 2091
fibroblast cells hybridized to NimbleGen ENCODE arrays.
Thanks to Paul Giresi at the Jason Leib lab for providing
these data.
-
NHGRI DNaseI-HS Raw (in progress) - DNase-chip raw
and p-value data in GM06990, CD4+, and Hela S3 cells.
Thanks to our UCSC staff who worked on these tracks:
Rachel Harte and Hiram Clawson (development), Ann Zweig
and Archana Thakkapallayil (quality review), and Donna
Karolchik (documentation).
The ENCODE data status page has been updated to reflect the
recent activity.
14 June 2006 - New ENCODE data at UCSC
During the build-up to the analysis paper submissions,
UCSC received a flurry of ENCODE data submissions
(6 during the month of May). We have recently
released three data sets to our public server;
the remaining tracks are in progress, as indicated below.
Released data:
-
Sanger ChIP-chip (HFL-1 cells) -
New data added to the Sanger ChIP track show the
location of modified histones in HFL-1 (embryonic lung
fibroblast) cells. Thanks to Rob Andrews and
Christopher Koch for providing this data.
-
DLESS (Detection of LinEage Specific Selection) -
This track shows elements predicted by the DLESS program
to be under lineage-specific selection, based on
alignments of 17 mammalian species from NHGRI/PSU
TBA ENCODE alignments. DLESS is based on a phylo-HMM
with states for neutrally evolving and conserved regions,
and for gains and losses on each branch of the tree.
Thanks to Adam Siepel of Cornell University, who
developed the DLESS program, generated the data, and
loaded the annotation track.
-
UW/Regulome Dnase/Array -
This track displays DNaseI sensitivity in GM06990 cells,
using the DNase/Array methodology. Dnase/Array is a novel
method for isolating DNA segments corresponding to
specific DNaseI cleavage events on individual nuclear
chromatin templates. Thanks to Scott Kuehn at the
University of Washington for providing these data.
Thanks to our UCSC staff who worked on these tracks:
Rachel Harte (development), Ann Zweig, Kayla Smith,
and Archana Thakkapallayil (quality review).
In quality review:
- UNC FAIRE Peaks (Update, 2091 fibroblasts)
In progress:
- University of Uppsala Chip/Chip (butyrate treatment in HepG2)
- UW/Regulome QCP Dnase HS (update and new cell lines -- 16 tissues)
- PECAN Alignments from EBI
The ENCODE data status page has been updated to reflect the
recent activity.
13 January 2006 - Release of October freeze data complete
All datasets submitted for the October 2005 ENCODE freeze
have now been released. The tracks with their release
dates are:
- 13 Jan. - Yale ChIP-chip (5 new transcription factors)
- 3 Jan. - University of Uppsala Chip/chip (data correction)
- 2 Jan. - Yale RNA (update)
- 29 Dec. - U Virginia DNA Replication Timing and Predicted Origins
- 28 Dec. - UT Austin STAGE
- 21 Dec. - Affy RNA and Transfrags (HeLa update)
- 22 Nov. - Sanger ChIP-chip (5 histone modifications)
- 19 Nov. - UC Davis ChIP-chip (Pol2, Myc) in HeLa
- 18 Nov. - U North Carolina FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements)
- 1 Nov. - Affy RNA and Transfrags (update)
- 1 Nov. - UT-Austin Ng ChIP-chip (E2F4, c-Myc factors)
- 1 Nov. - GIS PET RNA (update)
- 1 Nov. - Gencode Genes and Introns (update)
- 1 Nov. - NHGRI DNaseI HS (update and new GM06990)
- 27 Oct. - UCSD/LI Ng ChIP-chip (update and new TFs)
- 14 Oct. - GIS ChIP/PET (STAT1)
- 21 Sep. - Boston University ORChID (update)
Posted on 17 Oct. 2005 - ENCODE hg17 Genome Browser Released
The ENCODE browser for UCSC human genome assembly hg17
(NCBI Build 35) is now available at
http://genome.ucsc.edu/ENCODE/encode.hg17.html
and from the
Regions (hg17)
item in the left (blue bar) menu of the ENCODE portal.
All datasets originally submitted in hg17 coordinates for
the June data freeze were directly loaded; the remaining
data were coordinate-converted using the UCSC liftOver
process. A total of 351 data tables were loaded into our
database.
NOTE: Many of these tracks will be updated with new
data from the October ENCODE data freeze.
Many thanks to all the ENCODE consortium members who
contributed data for this release. We'd also like to
thank UCSC team members Kate Rosenbloom for portal and
track development and Jennifer Jackson, Kayla Smith, Ann
Zweig, and Bob Kuhn for quality assurance.
Posted on 15 July 2005 - Regulome DnaseI HS, Oxford Recombination, and MSA Data Released
The remaining datasets submitted for the June 2005 ENCODE
data freeze have now been released as annotation tracks in
the hg16 browser. These tracks and their release dates are:
- 15 Jul. - TBA23 EvoFold RNA Structure Predictions
- 14 Jul. - Regulome DNaseI HS and Sens
- 14 Jul. - Regulome Plate Q/A and Amplicons
- 14 Jul. - MSA Consensus Elements
- 14 Jul. - TBA alignments with Conservation
- 14 Jul. - Stanford MLAGAN Alignments and Conservation
- 13 Jul. - Oxford Recombination
Posted on 12 July 2005 - EGASP genes, RIKEN CAGE tags, and Stanford RTPCR, ChIP-chip and DNA Methylation data released
Several new ENCODE data sets have been released in the UCSC
Genome Browser.
EGASP Full, Partial, and Update: These three
gene prediction tracks are from the ENCODE Gene Annotation
Assessment Project (EGASP) Prediction Workshop 2005.
The EGASP Full track shows 20 sets of gene predictions
originally submitted for the workshop, covering all 44
ENCODE regions. THE EGASP Partial track shows eight sets of
gene precdictions that were submitted for the workshop, but do not cover all ENCODE regions. The EGASP Update track
shows updated versions of some of the submitted predictions.
Thanks to Julien Lagarde at IMIM for providing the EGASP
Full and EGASP Partial data sets. Thanks to Tyler Alioto of
IMIM (GeneID-U12 and SGP2-U12), Deyou Zheng of Yale (Yale
Pseudogenes), Sarah Djebali of Ecole Normale Supérieure
(Exogean), Jonathan Allen of TIGR/Univ. Maryland (Jigsaw)
and Mario Stanke of the University of Gottingen (Augustus)
for providing their EGASP Update gene sets.
RIKEN CAGE Predicted Gene Start Sites: This track
shows the numbers of 5' cap analysis gene expression (CAGE)
tags that map to the genome at specific locations. Areas in
which many tags map to the same region may indicate a
significant transcription start site. Thanks to Albin
Sandelin at RIKEN and the FANTOM (Functional Annotation
of Mouse) Consortium for providing these data.
Stanford RTPCR: This track displays absolute
transcript copy numbers for 136 genes and 12 negative
control intergenic regions, determined by real-time PCR in
HCT116 cells.
Stanford ChIP-chip, Stanford ChIP-chip Smoothed
Score: These tracks display raw scores and
sliding-window mean scores for regions bound by the Sp1
and Sp3 transcription factors in three cell lines (HCT116,
Jurkat, K562), assayed by chromatin immunoprecipitation and
hybridization to Nimblegen oligo tiling arrays. The
Smoothed Score track is provided solely for visualization
purposes -- the raw scores should be used for data analysis.
Stanford Methylation Digest: This track displays
experimentally determined regions of unmethylated
CpGs in eight cell lines (BE2C, CRL1690, HCT116, HT1080,
HepG2, JEG3, SNU182, and U87MG). Genomic DNA was digested
with a cocktail of six methyl-sensitive restriction enzymes,
size-selected, amplified, labeled, and hybridized to
Nimblegen oligo tiling arrays.
Thanks to Nathan Trinklein for providing the Stanford
datasets.
We'd like to acknowledge the work of the UCSC Genome
Bioinformatics team members who produced these tracks:
Kate Rosenbloom, Angie Hinrichs, and Hiram Clawson
(development), Ann Zweig, Bob Kuhn, Galt Barber, Rachel
Hart, and Ali Sultan-Quarrie (quality review), and Donna
Karolchik (documentation).
Posted on 6 July 2005 - NHGRI DIPs, HapMap SNPs, Sanger Assoc annotation tracks released
The first datasets in the ENCODE Variation group are now
available in the UCSC browser.
NHGRI Deletion/Insertion Polymorphisms: All human
trace data from NCBI's trace archive were aligned to the
genome and processed using the programs ssahaSNP and
ssahaDIP to detect deletion and insertion polymorphisms.
Thanks to Jim Mullikin at NHGRI for performing the analyses
and providing these data.
HapMap Allele Frequencies: This track shows allele
frequencies for the four HapMap populations in the ten
ENCODE regions that have been resequenced for variation
(manually selected regions m010, m013, and m014 and
randomly selected regions r112, r113, r123, r131, r213,
r232, and r321). These data were obtained from HapMap
public release #16c.1. Thanks to the International HapMap
Project for making this information available.
Sanger Genotype-Expression Association: This track
displays associations among gene expression data from the
60 unrelated Centre d'Etude du Polymorphisme Humain (CEPH)
individuals of the International HapMap Project with SNPs
genotyped by HapMap,
in eight ENCODE regions (m010, m013, m014 and r123, r131,
r213, r232, and r321). The CEPH population is composed
of Utah residents with ancestry from northern and western
Europe. The expression data were generated with the
Illumina platform at the Wellcome Trust Sanger Institute.
Thanks to Manolis Dermitzakis at the Sanger Institute for
providing these data.
We'd also like to acknowledge the UCSC ENCODE team members
who worked on these tracks: Heather Trumbower, Daryl Thomas,
and Angie Hinrichs (development), Galt Barber and Ali
Sultan-Qurraie (quality assurance), and Donna Karolchik
(documentation).
Posted on 23 June 2005 - Yale and Affymetrix ChIP-chip and transcription data release; New ENCODE track groupings
To aid ENCODE analysis and reduce visual clutter, we have
split the Genome Browser ENCODE track group into six new
groups:
- ENCODE Regions and Genes
- ENCODE Transcript Levels
- ENCODE Chromatin Immunoprecipitation
- ENCODE Chromosome, Chromatin and DNA Structure
- ENCODE Variation
- ENCODE Comparative Genomics
All of these track groups are visible on the UCSC test
browser. The last two groups, Variation and Comparative
Genomics, do not yet have published tracks on the public
server and therefore are not visible on that server.
We have also released a set of new Yale
data and an extensive update of Affymetrix data. The track
controls for these datasets can be found in the
track groups ENCODE Transcript Levels and ENCODE Chromatin
Immunoprecipitation.
Yale ChIP-chip and RNA: Three tracks of ChIP-chip
data from Yale, evaluating microarray platforms, have been
released: Yale ChIP pVal, Yale ChIP Sig, and Yale ChIP
Sites. These tracks show results of ChIP experiments using
STAT1 antibody in HeLa cells on four different microarrays
-- three custom maskless photolithographic oligo arrays,
designed at different resolutions, and the PCR amplicon
array developed by the Ren lab at the Ludwig Institute/UCSD.
Two tracks of RNA transcript data from Yale have been
released: Yale RNA and Yale TAR. These tracks show
transcriptionally active regions and transcribed
fragments for three cell types (neutrophil, placenta, and
NB4 variously treated for differentiation).
Thanks to Joel Rozowsky at Yale for providing this data.
Additional Yale ChIP-chip data is currently under review by
our quality assurance group.
Affymetrix ChIP-chip and RNA:
The Affymetrix ChIP-chip dataset now contains experimental
results for ten factors (Brg1, CEBPe, CTCF, H3K27me3,
H4Kac4, P300, PU1, Pol2, RARA, and SIRT1) in HeLa cells at
four timepoints after retinoic acid treatment, plus TFIIB
for the final timepoint only. The data is displayed in
eight tracks: Affy PVal 0h, 2h, 8h, 32h and Affy Sites 0h,
2h, 8h, 32h. We acknowledge that this track grouping
is a bit awkward and are working composite track
enhancements to provide more flexibility.
The Affy RNA tracks show RNA abundance and transfrags in
retinoic acid-stimulated HL-60 cells at four timepoints,
and in GM06990 and HeLa cells: Affy RNA Signal and Affy
Transfrags.
Thanks to Stefan Bekiranov and Srinka Ghosh at Affymetrix
for providing these data.
Thanks also to the UCSC ENCODE
team members who developed and reviewed these tracks and
to Rachel Harte in the UCSC browser group
for her assistance with track review.
Posted on 15 June 2005 - Six data sets released in Genome Browser
Six more ENCODE annotation tracks have been added to the
Genome Browser this week:
NHGRI DNaseI-Hypersensitive Sites (update): The
NHGRI DNaseI-HS track has been updated with new data. The
track now includes DNaseI-hypersensitive sites in CD4+
T-cells before and after activation by anti-CD3 and
anti-CD28 antibodies. Thanks to Greg Crawford at the
Collins lab (NHGRI) for providing these data.
Genome Institute of Singapore PET of PolyA+ RNA:
The GIS PET RNA track displays starts and ends of mRNA
transcripts determined by paired-end ditag sequencing in
two cell lines, MCF7 and HCT116 treated with 5
fluoro-uracil. A total of 584,624 PETs were generated for
MCF7 and 280,340 were generated for HCT116. More than
80% of the PETs in each group were mapped to the genome.
Thanks to Atif Shahab, Yijun Ruan, the GIS, and the
Bioinformatics Institute of Singapore for providing these
data.
Gencode Gene Annotations and Intron Validation:
The Gencode Genes track displays high-quality manual
annotations in the ENCODE regions generated by the
GENCODE project. A companion track, Gencode Introns,
shows experimental gene structure validations for these
annotations. Thanks to the HAVANA team at the Wellcome
Trust Sanger Institute; France Denoeud, Julien Lagarde,
and Roderic Guigo at the IMIM; and Alexandre Reymond
at the University of Geneva for providing the
annotations and experimental confirmation, as well as
working with UCSC to develop the track display.
Boston University Hydroxyl Radical Cleavage:
The BU ORChID track displays predicted hydroxyl radical
cleavage intensity on naked DNA for each nucleotide in
the ENCODE regions. The prediction algorithm draws data
from a database of 150 experimentally-determined cleavage
patterns. Thanks to Jay Greenbaum at the Tullius lab for
providing these data.
UC Davis ChIP-chip: The UCD Ng E2F1 track shows
ChIP analysis of HeLa cells using E2F1 antibody as
assayed on Nimblegen arrays. The E2F1 factor is associated
with cell cycle control, transcriptional regulation, and
apoptosis. Thanks to the Farnham lab and Kyle Munn, Todd
Richmond and Roland Green of Nimblegen for providing
these data.
Ludwig Institute/UCSD ChIP-chip:
Three tracks of ChIP-chip data using Nimblegen arrays
have been released: LI Ng TAF1 IMR90, LI Ng Validation,
and LI Ng gIF ChIP. The TAF1 track shows genome-wide
TAF1 binding sites as determined by ChIP in IMR90
(fibroblast) cells assayed on Nimblegen high-density
oligo arrays. The peaks from the genome scan experiments
were verified using four antibodies associated with
transcription start (Pol2, H3ac, H3K4me2, and TAF1) on
condensed arrays covering the putative TAF1 binding sites.
The gIF track shows the results of ChIP-chip experiments
using gamma interferon-treated HeLa cells, with Pol2 and
H3K4me3 antibodies.
Two additional tracks, LI ChIP Various and LI gIF ChIP,
display ChIP-chip data assayed on Ren lab ENCODE PCR
tiling arrays for five antibodies (Pol2, TAF1, H3K4me2,
SUZ12, H3K27me3) and four cell lines (HeLa, THP1, IMR90,
HCT116), as well as gamma interferon experiments using
seven antibodies (Pol2, TAF1, H3K4me2, H3K4me3, H3ac,
H4ac, and STAT1) in HeLa cells.
Thanks to Chunxu Qu and Bing Ren, at the Ren lab for
providing these data and assisting with track display
issues.
We'd also like to acknowledge the UCSC team members who
developed these tracks: Kate Rosenbloom, Hiram Clawson,
and Angie Hinrichs for track development; Bob Kuhn, Ali
Sultan-Qurraie and Galt Barber for quality assurance;
and Donna Karolchik and Jim Kent for documentation.
Posted on 9 June 2005 - Sanger Histone Modification ChIP-chip track updated
The Sanger Institute has submitted ChIP-chip data for
additional antibodies and cell lines, which we have
incorporated into the existing Sanger ChIP browser track.
The expanded track now contains data for five antibodies
(H3K4me1, H3K4me2, H3K4me3, H3ac, H4ac) and two cell
lines (GM06990, K562 (leukemia)).
Thanks to Rob Andrews and Chris Koch at the Dunham lab
for providing these data. UCSC team members who developed
this track include Hiram Clawson (track development), Ali
Sultan-Qurraie (quality assurance), and Donna Karolchik
and Jim Kent (documentation).
Posted on 6 June 2005 - Changes to ENCODE track labels in Genome Browser
We have modified the Genome Browser labels for the
existing ENCODE data tracks to trim overly-long labels
that were truncated in the display and to facilitate
cross-track analysis. The new label format shows the
submitter and the experiment, followed by the cell line
(in tracks where the data includes only one cell line).
Posted on 6 June 2005 - Stanford Promoters and UVa DNA Replication tracks released
We are pleased to announce the first UCSC Genome Browser
tracks released for the June 2005 ENCODE data freeze:
Stanford Promoters and UVa DNA Replication Temporal
Profiling.
Stanford has provided an update of their promoter activity
data based on transient transfection luciferase reporter
assays of 643 putative promoter fragments in the ENCODE
regions. The update includes two additional cell lines
and activity averaged across all cell lines. The data
tables now contain additional experimental detail to
facilitate analysis. This track, containing 17 subtracks
(16 cell lines and the average), is labeled "Stanf. Promoter". Thanks to Sara Hartman at
the Myers lab for providing these data.
The Dutta lab at Univerity of Virginia (UVa) has completed
the second biological replicate of their temporal
profiling of HeLa cell replication products and has
provided a dataset containing merged data from the two
replicates. The track, containing five subtracks
representing two-hour intervals, is labeled
"UVa DNA Rep". Thanks to Christopher Taylor at
the Dutta lab for providing these data.
We'd also like to acknowledge the UCSC team members who
worked on these annotation tracks: Angie Hinrichs (track
development), Galt Barber and Ali Sultan-Qurraie (QA), and
Jim Kent and Donna Karolchik (track documentation).
Posted on 24 May 2005 - New MSA sequence data freeze available
A new ENCODE MSA sequence data freeze is available on the
UCSC downloads server. The latest freeze contains
sequences from 23 vertebrates provided by NISC, Baylor,
the Broad Institute (2X) and the whole genome shotgun
(WGS) assemblies. The data may be downloaded as
individual data files or a
directory tarball.
Aligners are encouraged to upload alignments and related
data (such as conservation scores and elements) to the
UCSC ENCODE ftp site as soon as possible and then notify
Kate Rosenbloom.
Other data, (conservation, trees, etc.)
will be generated based on this dataset.
The following is a summary of data updates from the
previous release:
-
The human assembly version remains at hg16 (Jul. 2003).
-
The mouse assembly has been updated from mm5 (May 2004) to
mm6 (Mar. 2005).
-
The multiple rat sequences have been replaced by a single
sequence: rn3 (Jun. 2003).
-
The cow sequences have been updated using an assembly of
BAC-based sequences provided by Baylor College of
Medicine.
-
Fugu (fr1), macaque (rheMac1), opossum (monDom1),
Tetraodon (tetNig1), Xenopus (xenTro1), and zebrafish
(danRer2) have been added. The macaque sequence was
obtained from a Baylor
College of Medicine assembly that has not yet been
officially released.
-
A new NISC species, rfbat
(Rhinolophus ferrumequinum), is now available.
-
Platypus data from regions
where NISC has not yet generated data were provided by
Jim Mullikin from a preliminary assembly of Washington
University WGS reads.
-
This freeze includes low-redundancy sequence data from
tenrec, elephant, armadillo, and rabbit. Only one set of
sequences are provided per species/target combination;
where available, NISC data is provided instead of
the 2X assemblies. These data are not yet accessioned at
NCBI, but were made available by the Broad Institute
(rabbit, elephant, armadillo) and Jim Mullikin (tenrec).
-
Orthology predictions for Fugu were made only by
MAVID/Mercator; predictions for all other assemblies
supported by the UCSC Genome Browser represent a union
with UCSC predictions as well. Because no additional
post-processing was done on the Fugu predictions, they
contain a few very small contigs.
Thanks to the many people, particularly Elliott Margulies
and Daryl Thomas, who made this release possible.
Posted on 24 May 2005 - ChIP-PET/GIS annotation track (Genome Institute of Singapore) released
The ENCODE Genome Browser now features the ChIP-PET/GIS
annotation track, which shows paired-end
ditag (PET) sequences derived from 65,572 individual p53
ChIP fragments of 5-fluorouracil (5FU) stimulated HCT116
(colon) cells. Only PETs with a single specific mapping
to the genome are included in this track.
Thanks to Atif Shahab, Chia-Lin Wei, and Yijun Ruan at the
Genome Institute of Singapore for
providing the p53 ChIP-PET library and sequence data. The
data were mapped and analyzed by scientists from the
Genome Institute of Singapore, the Bioinformatics
Institute, Singapore, and Boston University. For more
information about this annotation, see the ChIP-PET/GIS
track description page.
Posted on 23 May 2005 - Boston University First Exon annotation track released
The First Exon/BU annotation track, contributed by the
ZLAB at Boston University, is now
available in the UCSC Genome Browser. This track displays
expression levels of computationally identified first
exons and a constitutive exon of 20 genes in the ENCODE
regions.
For each gene, all alternative first exons were identified
based on manual selection of predictions from the
PromoSer
program. The expression levels of exons were then
quantified using rcPCR in ten normal human tissues.
Thanks to Ulas Karaoz and the Zhiping Weng lab at
Boston University for providing these data. For more
information about this annotation, as well as a complete
list of the individuals who contributed to this track,
see the First Exon/BU track description page.
Posted on 7 May 2005 - ENCODE status page now available
A simple summary page has been added to the UCSC ENCODE
portal to show the status of datasets submitted to UCSC by
ENCODE contributors. The page may be found at
http://genome.ucsc.edu/ENCODE/trackStatus.html and can be accessed via
links on the ENCODE home page and the ENCODE data
submission page. The status page will be updated
approximately once a week.
To review the latest ENCODE data releases, see the
release log.
Posted on 6 May 2005 - UCSD/Ludwig Institute histone modification ChIp/Chip data released
Two additional sets of ChIp/Chip data from UCSD/Ludwig
Institute are now available:
ChIp/LI AcH3
ChIp/LI MeH3K4
These tracks display locations of acetylated H3 and
dimethylated K4H3 binding in IMR90 (lung fibroblast)
cells.
To consolidate viewing in the browser, the previously
released eight datasets from UCSD/LI have been reformatted
as two composite tracks (one track per antibody) with
each track containing four subtracks (one per cell line).
These tracks are:
ChIp/LI Pol2
ChIp/LI TAF1
To facilitate data analysis, the data were also reloaded
in a format that allows extraction of the original data
values via the UCSC table browser.
Thanks to Chunxu Qu and the
Ren
lab for providing these data.
Posted on 17 Feb. 2005 - ENCODE downloads released
The ENCODE download area has been reorganized and updated
on our public download server. The downloads access page
is now:
http://hgdownload.cse.ucsc.edu/goldenPath/encode/
and the annotations are now located in the
assembly-specific download directory, currently:
http://hgdownload.cse.ucsc.edu/goldenPath/hg16/encode/
Any web pages referencing the previous UCSC ENCODE
downloads will need to be updated. Please contact us if
you have any difficulties.
Posted on 2 Feb. 2005 - Affymetrix ChIp/Chip and PolyA RNA data released
A second set of ENCODE ChIp/Chip data is now available on
the July 2003 human genome assembly:
ChIp/Affy Pol2 Pval
ChIp/Affy Pol2 Sites
RNA/Affy Signal
RNA/Affy Sites
These tracks show RNA Polymerase II precipitation and
RNA abundance in retinoic acid-stimulated HL-60 cells at
0, 2, 8, and 32 hours, as measured by Affymetrix tiling
arrays in the non-repetitive ENCODE regions. The
Pval and Signal tracks show values
for each tiled probe; the Sites tracks show
contiguous regions of enrichment.
A new composite track display was developed to
concisely display multiple data sets of similar types, a
common feature of ENCODE data. Each of these new tracks
contains 4 subtracks, one for each time interval. The
subtracks share a single description page and set of
visibility controls. Checkboxes on the track
configuration page allow selected subtracks to be hidden
in the display.
These data were generated and analyzed by Tom Gingeras'
group at
Affymetrix and
Kevin Struhl's group at Harvard Medical
School. We would like to thank Stefan Bekiranov at
Affymetrix for submitting the data and working closely
with us to clarify the experimental methods and
verification descriptions.
Posted on 5 Nov. 2004 - First ENCODE data tracks in the UCSC Browser
The first datasets submitted for the ENCODE project are
now publicly available:
-
ChIP-chip and transcription (Ludwig Institute/UCSD)
-
Temporal profiling of DNA replication (University of
Virginia)
-
Promoter activity (Stanford)
-
DNaseI hypersensitive sites (NHGRI)
These tracks are visible in the ENCODE track group of the
July 2003 (hg16) human genome assembly. We would like to
thank the labs of Bing Ren (LI/UCSD), Anindya Dutta (UVA),
Rick Myers (Stanford), and Francis Collins (NHGRI) for
contributing the initial ENCODE data sets.
Posted on 26 Oct. 2004 - Sequence Freeze For Multiple Alignments
We are pleased to release the first "official" sequence
data freeze for the ENCODE multiple sequence alignment
projects. The data formats are described in the
README file, and the sequences and
supporting information is collected in the
data directory.
The species included in this freeze are as follows:
_SPECIES_ _SOURCE_
Human hg16
Chimpanzee panTro1
Dog canFam1
Rat rn3
RatB BCM
Mouse mm5
Chicken galGal2
Galago NISC
Baboon NISC
Marmoset NISC
Armadillo NISC
Platypus NISC
You will notice that we have worked hard to include a
number of species for which genome-wide sequence data was
already available. The process by which these orthologous
regions were identified is still an area of active
research development, the details of which will be
presented at the upcoming ENCODE meeting at CSHL.
Please note that we have also included a *second* rat
sequence (ratB) in this freeze. RatB represents an
initiative to standardize the quality level of sequences
in the ENCODE regions for species with genome-wide
sequence data. The data were made available just before
our freeze date, so we decided to include both versions of
the rat sequence for now. Eventually these sequences will
likely be rolled into future genome assemblies.
Remember, this is a work in progress, so not all targets
have sequence from all species. And some species/target
combinations may not be complete yet. Progress on the
NISC-generated sequences can be found at the
NISC ENCODE Project: Comparative
Sequencing.
Many people have worked very hard to make these data
available. Special thanks to Daryl Thomas, Kate
Rosenbloom and the entire UCSC Team; Greg Schuler and the
NCBI team; Colin Dewey and Lior Pachter; Pam Thomas and
the NISC team; and David Wheeler and the BCM team.
Please send any comments to the
ENCODE MSA Mailing List.
Elliott
--
Elliott H. Margulies, Ph.D.
Genome Technology Branch, NHGRI
Posted on 24 Jun. 2004 - ENCODE Project Portal Released
We are proud to announce the release of features
in the UCSC Genome Browser that are tailored to
the ENCODE project community, including this home
page to consolidate these resources.
The initial resources include sequences for the current
human assemblies (hg16, hg15, hg13, and hg12), sequence of
the comparative species from
NISC,
tools for coordinate conversion between human
assemblies, format descriptions for data
submission, and contact information for help with
submitting annotation data and analyses.
Bulk downloads of the sequence and annotations may
be obtained from the ENCODE Project
Downloads
page. The sequences available here are repeat-masked
versions of the GenBank records.
| 


